Intel® oneAPI Data Analytics Library Developer Guide and Reference
A newer version of this document is available. Customers should click here to go to the newest version.
Visible to Intel only — GUID: GUID-452F2D3C-69A7-4895-B912-20E1AF77F187
Visible to Intel only — GUID: GUID-452F2D3C-69A7-4895-B912-20E1AF77F187
Distributed Processing
This mode assumes that the data set is split into nblocks blocks across computation nodes.
Algorithm Parameters
The K-Means clustering algorithm in the distributed processing mode has the following parameters:
Parameter |
Default Value |
Description |
|---|---|---|
computeStep |
Not applicable |
The parameter required to initialize the algorithm. Can be:
|
algorithmFPType |
float |
The floating-point type that the algorithm uses for intermediate computations. Can be float or double. |
method |
defaultDense |
Available computation methods for K-Means clustering:
|
nClusters |
Not applicable |
The number of clusters. Required to initialize the algorithm. |
gamma |
1.0 |
The weight to be used in distance calculation for binary categorical features. |
distanceType |
euclidean |
The measure of closeness between points (observations) being clustered. The only distance type supported so far is the Euclidean distance. |
assignFlag |
false |
A flag that enables computation of assignments, that is, assigning cluster indices to respective observations. |
To compute K-Means clustering in the distributed processing mode, use the general schema described in Algorithms as follows:
Step 1 - on Local Nodes
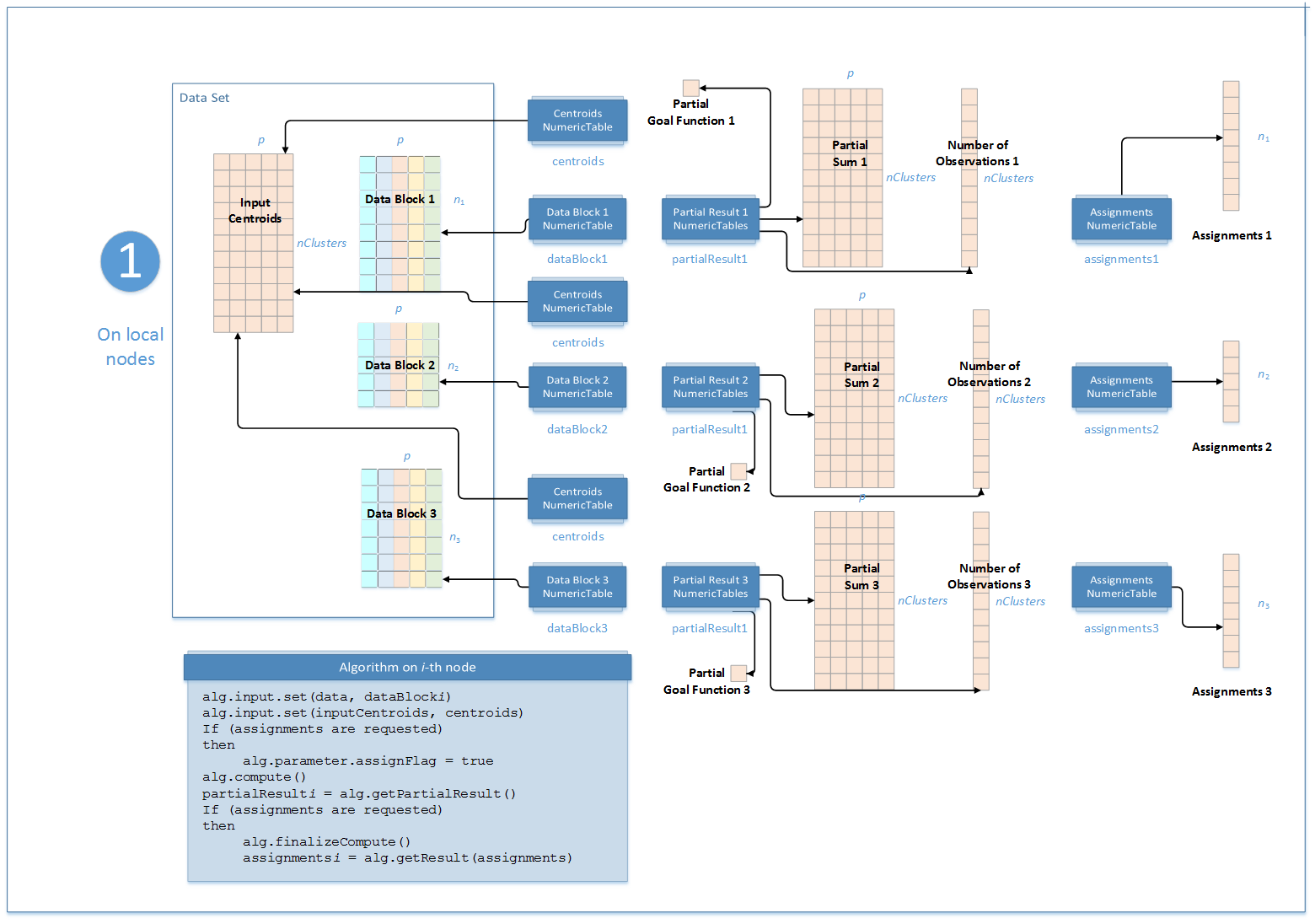
In this step, the K-Means clustering algorithm accepts the input described below. Pass the Input ID as a parameter to the methods that provide input for your algorithm. For more details, see Algorithms.
Input ID |
Input |
|---|---|
data |
Pointer to the |
inputCentroids |
Pointer to the |
In this step, the K-Means clustering algorithm calculates the partial results and results described below. Pass the Partial Result ID or Result ID as a parameter to the methods that access the results of your algorithm. For more details, see Algorithms.
Partial Result ID |
Result |
|---|---|
nObservations |
Pointer to the
NOTE:
By default, this result is an object of the HomogenNumericTable class, but you can define this result as an object of any class derived from NumericTable except CSRNumericTable.
|
partialSums |
Pointer to the
NOTE:
By default, this result is an object of the HomogenNumericTable class, but you can define the result as an object of any class derived from NumericTable except PackedTriangularMatrix, PackedSymmetricMatrix, and CSRNumericTable.
|
partialObjectiveFunction |
Pointer to the
NOTE:
By default, this result is an object of the HomogenNumericTable class, but you can define this result as an object of any class derived from NumericTable except CSRNumericTable.
|
partialCandidatesDistances |
Pointer to the
NOTE:
By default, this result if an object of the HomogenNumericTable class, but you can define this result as an object of any class derived from NumericTable except PackedTriangularMatrix, PackedSymmetricMatrix, CSRNumericTable.
|
partialCandidatesCentroids |
Pointer to the
NOTE:
By default, this result if an object of the HomogenNumericTable class, but you can define this result as an object of any class derived from NumericTable except PackedTriangularMatrix, PackedSymmetricMatrix, CSRNumericTable.
|
Result ID |
Result |
|---|---|
assignments |
Use when assignFlag = true. Pointer to the
NOTE:
By default, this result is an object of the HomogenNumericTable class, but you can define this result as an object of any class derived from NumericTable except PackedTriangularMatrix, PackedSymmetricMatrix, and CSRNumericTable.
|
Step 2 - on Master Node
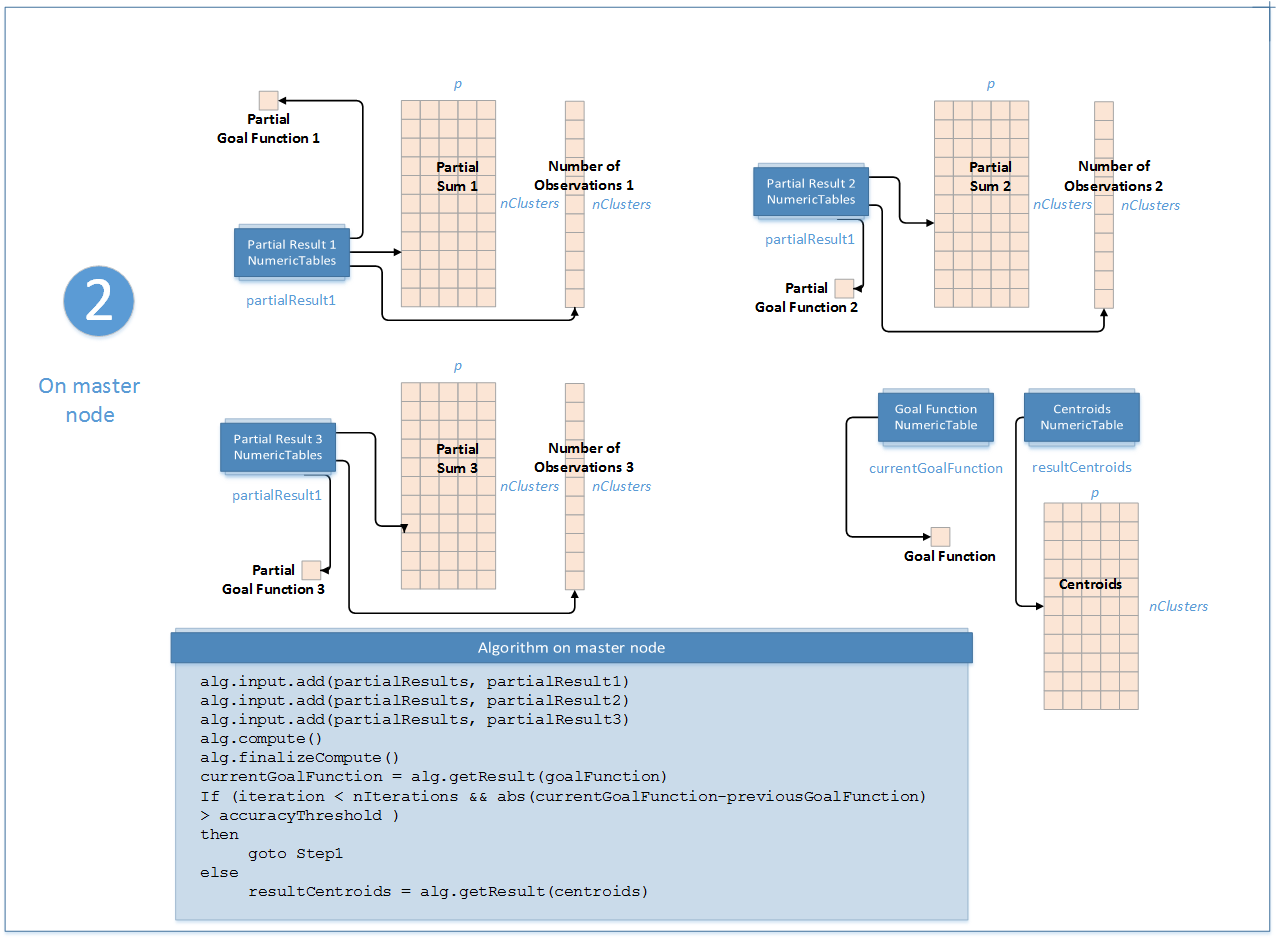
In this step, the K-Means clustering algorithm accepts the input from each local node described below. Pass the Input ID as a parameter to the methods that provide input for your algorithm. For more details, see Algorithms.
Input ID |
Input |
|---|---|
partialResuts |
A collection that contains results computed in Step 1 on local nodes. |
In this step, the K-Means clustering algorithm calculates the results described below. Pass the Result ID as a parameter to the methods that access the results of your algorithm. For more details, see Algorithms.
Result ID |
Result |
|---|---|
centroids |
Pointer to the
NOTE:
By default, this result is an object of the HomogenNumericTable class, but you can define the result as an object of any class derived from NumericTable except PackedTriangularMatrix, PackedSymmetricMatrix, and CSRNumericTable.
|
objectiveFunction |
Pointer to the
NOTE:
By default, this result is an object of the HomogenNumericTable class, but you can define this result as an object of any class derived from NumericTable except CSRNumericTable.
|