Introduction
Computational fluid dynamics (CFD) based blood flow simulations using image-derived vascular geometries have demonstrated potential in cardiovascular disease evaluation and management. Researchers in the group led by Professor Amanda Randles developed HARVEY—a massively parallel CFD code capable of resolving patient-derived blood flow at cellular resolutions in complex vascular geometries (Figure 1). HARVEY has been employed for the evaluation of cardiovascular disease through noninvasive determination of patient biomarkers.
Originally, the HARVEY GPU code was written to run only on NVIDIA* devices. To take advantage of the compute capability of upcoming exascale machines such as Aurora, powered by the Intel® Data Center GPU Max Series, we ported the code to run across multiple GPU node architectures.
Intel® toolkits were used to port the original HARVEY code from CUDA* to Data Parallel C++ (DPC++) and optimized for Intel Data Center GPU Max Series on the Aurora testbed system, Sunspot, that is maintained by Argonne National Laboratory. With the help of Intel® oneAPI profiling tools, investigators were able to achieve greater than 10x performance improvement compared to baseline.

Figure 1. Pulsatile flow in an image-derived geometry of a human aorta performed using HARVEY.
Image Source: Joe Insley, Argonne National Laboratory.
oneAPI
oneAPI is an industry-wide effort based on an open standards specification. It provides cross-architecture, cross-vendor portability while providing great performance. oneAPI is a foundational programming stack and is used to optimize middleware and frameworks that sit on top of it.
Porting HARVEY with oneAPI
Using the Intel® DPC++ Compatibility Tool, the vast majority of HARVEY code was automatically ported to DPC++. This saved us months of code engineering, enabling us to get up and running quickly with an initial working code capable of running both on Intel GPUs on the Sunspot testbed, as well as on NVIDIA* A100 GPUs on the Polaris supercomputer (Argonne National Laboratory). The overall study is depicted in Figure 2.

Figure 2. Study overview in which HARVEY was ported from CUDA to DPC++, as well as HIP and Kokkos and run on Sunspot (Intel® Data Center GPU Max Series), Polaris (NVIDIA A100 GPU), Summit (NVIDIA V100 GPU), and Crusher (AMD* MI250X GPU).
Reference: Martin, Aristotle, et al. "Performance Evaluation of Heterogeneous GPU Programming Frameworks for Hemodynamic Simulations." Proceedings of the SC'23 Workshops of The International Conference on High Performance Computing, Network, Storage, and Analysis, 2023.
Removing Bottlenecks
Intel® VTune™ Profiler, a component of the oneAPI software ecosystem, is an advanced performance-analysis tool used to assist developers with optimizing application performance. Using Intel VTune Profiler, we identified that host-device memory transfers were taking a significant fraction of the iteration time.

We lifted this bottleneck by offloading memory-transfer-intensive boundary condition computations onto the GPU. The updated Intel VTune Profiler results indicated that we were better using the GPUs with time spent in the main computational kernel being dominant over host-device transfers.

These improvements in turn resulted in more than 10x speedup on Intel GPUs on Sunspot.
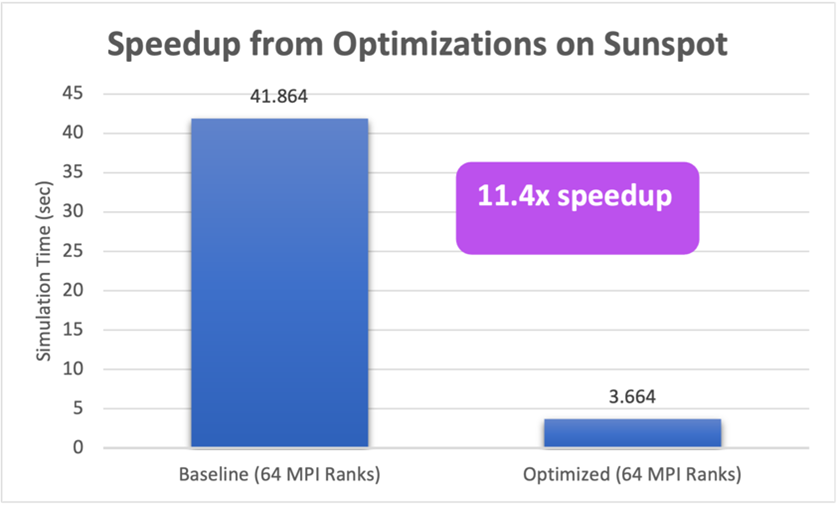
In the preceding chart, we ran a benchmark simulation with HARVEY on 64 Intel GPU tiles (with one MPI rank per tile). In one case, we ran our original ported SYCL* code, which gave a simulation time of 41.864 seconds, and in the other case, we ran the code with the profiler-guided optimizations applied, which produced the same results in 3.664 sec, yielding an order of magnitude speedup.
"Working with the Intel toolkit provided a very straightforward means to getting our code up and running on the Intel GPUs. It allowed Aristotle, the graduate student leading the work, to quickly get not only a proxy app but the full codebase ported over, which was critical in getting the at-scale performance data for our P3HPC paper."
Visualization Using the Intel® Rendering Toolkit
Scientific visualization, broadly speaking, involves taking numbers that are generated through large and complex simulations and presenting them as plots, graphs, or pictures. Scientific visualization plays an important role in the analysis of a wide variety of data from fields like astronomy, physics, and biomedical science. These types of visualizations give researchers and scientists insight into the data and help them extract more value from the simulations, sticking true to the old saying of a picture being worth a thousand words or, in this case, numbers. To be able to visualize such large and complex data on different kinds of devices, it is important for the visualization algorithm to be efficient, scalable, and portable.
HARVEY’s visualization requirements demonstrate the demand for high-performance, high-fidelity rendering capabilities such as the Intel® Rendering Toolkit (Render Kit). Render Kit is a collection of fully open source, multiplatform libraries that provide a feature set to address a myriad of unique rendering requirements. Intel® Embree provides routines for high-performance ray-surface intersection queries, Intel® Open Volume Kernel Library (Intel® Open VKL) focuses on providing efficient solutions for volume intersection queries. Intel® Open Image Denoise is an AI-optimized denoiser that can be used to remove any resulting noise to produce high-quality images. Intel® OSPRay is a scalable rendering framework that builds on top of these libraries and can be used for ray tracing, path tracing, and volume rendering. Multiple applications can use Intel OSPRay, such as our own software Intel® OSPRay Studio, or third-party applications such as ParaView*, VisIt, and SENSEI*.
HARVEY’s blood flow simulation data is large and complex enough that traditional visualization methods would not provide the researchers with the level of interactivity they would have liked. Intel OSPRay is widely used in the scientific visualization community because it provides scalable rendering through its MPI module. Intel OSPRay supports two methods of parallel rendering. The first method is image parallel. In this method, the image workload is distributed across different nodes and the data is replicated. The second method is data parallel. In this method, the data is distributed across different nodes. The data parallel method is used when the size of the data is too large to fit into the memory of a single node. Both modules enable efficient rendering for large workloads by distributing the computation across multiple MPI instances. The data generated by the HARVEY code is larger than the memory of a single compute node, and using the data parallel method enables the visualization of such high-resolution data over multiple time steps.
Support for Intel GPUs was added through SYCL with the release of Intel OSPRay 3.0, bringing the uniqueness of Intel OSPRay with its scalable rendering framework via MPI to powerful GPU hardware. Intel OSPRay integration within multiple third-party applications was also enhanced to support Intel GPUs. One of these applications is the Kitware* ParaView* visualization tool. While the integration of Intel OSPRay within ParaView is in the process of being made official, it has been used to visualize simulation data generated by the HARVEY code on 96 GPUs across 16 nodes on Sunspot. The data that was visualized consisted of 100 time steps using 900 GB on disk and resulted in a peak memory usage of 80 GB of VRAM per GPU. Each time step consists of rank-partitioned surface and volumetric data in a VTK file format, the different data partitions are distributed across multiple MPI instances. ParaView uses two components when rendering on HPC clusters: a client that runs on the local machine and a server that runs on the supercomputer. The user can interact with the visualization via the ParaView client. An example of this is presented in Figure 3 where the visualization was rendered on 96 GPUs on Aurora Sunspot, while the ParaView client was running on a local machine. ParaView, also provides a batch mode, where the ParaView server runs the visualization script without connecting to a client on the local machine and the rendered images are saved. The visualization in Figure 4 was rendered using this mode. The visualizations show the computed trajectories and deformations of red blood cells and a larger cancerous cell as they travel through blood vessels.

Figure 3. Visualizing tumor cell trajectories on 16 nodes on Sunspot using Intel Data Center GPU Max Series. The visualization tool in the image is Kitware ParaView and it uses Intel OSPRay to render the image. In this image, ParaView is running in client/server mode, where the visualization routine is running in real time. Dataset: HARVEY, Amanda Randles, Duke University, and Oak Ridge National Laboratory. Visualization: David DeMarle, Intel.

Figure 4. Visualizing tumor cell trajectories on Sunspot. The image is rendered with Intel OSPRay on Intel Data Center GPU Max Series using Kitware ParaView and produced using batch mode. Dataset: HARVEY, Amanda Randles, Duke University, and Oak Ridge National Laboratory. Visualization: Joseph Insley, Argonne National Laboratory's Leadership Computing Facility.
Conclusion
Intel® Toolkits provides users with probability and high performance. The Intel DPC++ Compatibility Tool efficiently helps users in the HPC community prepare large-scale scientific and engineering codes for production runs on cross-architecture platforms with minimal porting time and profiling analysis, facilitating architecture-specific optimizations. The Render Kit generates high-quality images that can be used to gain a better understanding of the simulation data, which leads to scientific and engineering advancement.
The next step will be porting the HARVEY cell code using the Intel DPC++ Compatibility Tool, enabling it to use the full exascale power of the Aurora supercomputer to run fluid-structure interaction simulations of red blood cells in arterial geometries, and the Render Kit to enable visualization of these massively parallelized simulations.