Scalability, Performance, and Flexibility
Venkat Krishnamurthy, vice president of product management, OmniSci*
@IntelDevTools
Get the Latest on All Things CODE
Sign Up
It’s no secret that over the last ten years, the landscape of technology has been driven forward primarily by data (see Data and AI Landscape 2019). In every sphere of human endeavor, we now collect and use data as a primary way to solve problems—whether it’s for pure and applied sciences or industry. As a result, data science has emerged as an important discipline in its own right. Being able to organize and derive useful insights from ever-growing datasets is a crucial skill set. A whole universe of tools and techniques has emerged to enable data scientists to ask bigger and deeper questions of data.
The typical workflow for a data scientist is basically an iterative process (Figure 1).

Figure 1. The data science workflow
Data scientists have long favored the Python* and R* ecosystems for a combination of ease of use (for data scientists, not professional programmers) and the extensive supporting ecosystem of libraries for statistical and numerical computing. Lately, the emergence of deep learning and AI as critical subfields in data science has pushed even more capabilities into these ecosystems. Python in particular enjoys widespread popularity for machine-learning and AI workflows.
In the Python world, the PyData stack (Figure 2) has been the most complete and popular set of data science tools for some time. Starting from the lowest level with numeric computing on N-dimensional array data (NumPy), this stack provides successive layers for scientific computing (SciPy), tabular and relational data analysis (pandas), and symbolic computing (SymPy).
Further up the stack, there are specialized libraries for visualization (Matplotlib, Altair*, and Bokeh), machine learning (scikit-learn*), and graph analytics (NetworkX and others). Domain-specific toolkits such as Astropy and Biopython, in turn, build on these layers, providing a deep and rich ecosystem of open tools. Alongside these resources, Project Jupyter* is a significant revolution driving the idea of interactive computing in general, and data-driven storytelling in particular. Many data scientists use Jupyter as their default development environment to create and explore hypotheses and models.

Figure 2. The Python data science (PyData) stack
OmniSci: Accelerated Analytics Exploiting Modern Hardware
At OmniSci, formerly MapD, we’ve pioneered using high-performance computing (HPC) techniques to simultaneously accelerate analytic SQL and data visualization since 2013. The open-source OmniSciDB SQL engine brings together multiple ideas in this regard:
- I/O acceleration through efficient use of the memory hierarchy
- LLVM-based just-in-time (JIT) compilation for analytic SQL kernels
- Large-scale in situ data visualization
- Efficient data interchange with out-of-core workflows such as machine learning and deep learning
It allows us to accelerate analytical SQL queries by two to three orders of magnitude, both on hardware accelerators like GPUs as well as on modern-vector CPUs.
While OmniSci initially focused on the problem of analytical SQL and data visualization (we open-sourced the OmniSciDB engine in 2017), we quickly learned we could provide deeper value to the open PyData ecosystem with some key integrations and interfaces. We wanted to ensure that we gave data scientists a programmatic workflow within the PyData stack—that is, enabling them to work inside Jupyter and perform operations on data using a familiar API (rather than just raw SQL). With this in mind, we worked with Quansight Labs*, founded by Travis Oliphant, the creator of NumPy and SciPy, on realizing an open-data science stack that used the scalability and performance of the core OmniSciDB engine with a well-layered set of Python APIs.
Dataframes and Related Problems
Along the way, we encountered some key problems with basic data science workflows that are worth highlighting due to their relation to analytical data structures. Comparable to tensors–the multidimensional arrays of data that are the primary data structure in machine learning and deep learning–the dataframe (Figure 3) is perhaps the most commonly used data structure in analytics because of how closely it maps to the existing relational database paradigm of a table.
Data scientists create dataframes from any sort of tabular data. The dataframe libraries exist in Python, with pandas being by far the most popular, R (where they originated), as well as newer languages like Julia*.

Figure 3. The structure of a dataframe
The problem is that as the number of analytics tools proliferated across ecosystems, the number of dataframe implementations grew as well. For example, Apache Spark* is a popular analytical processing engine and platform. It has a dataframe API (a distributed dataset of Spark) that provides many dataframe capabilities. But this is different from pandas, which is different from R. At OmniSci, we learned that all of these APIs, while incredibly flexible, suffer from a combination of scalability and performance issues that make it difficult for data scientists to truly and interactively explore large datasets.
For example, Spark allows large distributed dataframes, but the computational engine doesn’t allow for truly interactive (subsecond) querying because Spark was structured to be a distributed system first and foremost (and there are additional overheads from running on the Java virtual machine [JVM]). On the other hand, pandas is a rich, powerful API, but it suffers from scalability issues because the operations are implemented in Python and incur significant overhead from being run in an interpreted environment.
Finally, there are well-known interchange problems across languages and ecosystems (JVM, Python, R) that make it both difficult and inefficient to assemble complete workflows that can span different computational environments. For example, Spark is often used in the early stages of a workflow to manipulate and shape large datasets for further analysis, often in local computing environments like Python and pandas. Until recently, this incurred interchange and translation overhead limited scalability. Fortunately, Apache Arrow* has emerged as a standard to address this need. But it’s not pervasively used yet for data interchange between different frameworks.
Early in our data science journey, OmniSci invested in Ibis (Figure 4). A relatively less-known API (from the creator of pandas, Wes McKinney), Ibis provides an interesting path to bring large data processing and storage systems such as OmniSci into the Python data science stack. Ibis does this by providing several key layers that bridge the PyData stack into the world of data stores. According to the Ibis project website, these components include:
- A pandas-like, domain-specific language (DSL) designed specifically for analytics (that is, Ibis expressions) that enables composable, reusable analytics on structured data. If you can express something with a SQL SELECT query, you can write it with Ibis.
- Integrated user interfaces to Apache Hadoop* Distributed File System (HDFS) and other storage systems.
- An extensible translator-compiler system that targets multiple SQL systems.

Figure 4. Ibis, an API bridging data science and data stores
Supported SQL systems include MySQL*, PostgresSQL, BigQuery8 enterprise data warehouse, OmniSci, Spark, ClickHouse*, and others, and it’s easy to add a new back end.
We also invested in open-data visualization built on Altair*, a declarative Python visualization framework. Altair builds on the Vega and Vega-Lite modern, declarative user interface frameworks for data visualization. We worked with Quansight to unify Ibis and Altair so that visually exploring much larger datasets are now possible without moving data from the source systems.
OmniSci and Intel: Better Together for Data Science
In 2019, we released our first version of these integrations—again, all open sourced within the respective projects. At the time, we entered into what has been a fruitful collaboration with Intel, which saw the power and performance of the open-source OmniSciDB engine. Together, we’re exploring ways to bridge its capabilities into data science tooling (identical goals to us) to help accelerate dataframe-centric analytical workflows. The Intel team selected OmniSci as a reference platform to showcase the power of its ecosystem. The Intel team, with its deep systems and tuning expertise, has contributed in major ways to this goal.
Using profiling tools from Intel like Intel® VTune Profiler, the team identified several areas for performance tuning and optimization for running OmniSciDB (including the use of Intel® oneAPI Threading Building Blocks and better locking) on Intel® CPU families. OmniSci significantly outperforms a Spark cluster in all four queries of the Billion Row Taxi benchmark (Figure 5).
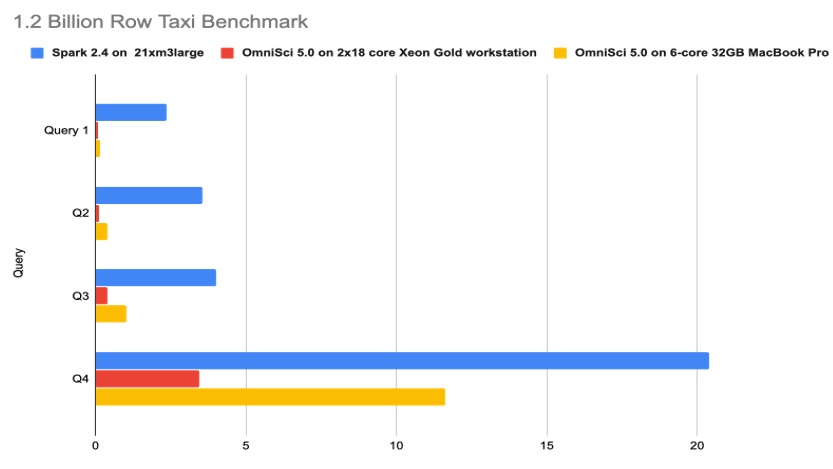
Figure 5. 1.2 Billion Row Taxi benchmark with OmniSci on Intel CPUs. The x-axis shows the time (in seconds) to complete each query. Get details on this benchmark.
The team also contributed a high-performance, ingestion component based on Arrow that can be used for in-memory data ingestion into OmniSci, bypassing the need to import data first. The goal is to support a workflow similar to how dataframes are created from static files like CSV or Apache Parquet*, but without having to use the OmniSci storage system underneath.
Work is now underway to componentize the OmniSciDB engine into a library, which facilitates embedding it in a data science workflow that's based on Python while taking advantage of the scale and performance of the engine. Work is also underway to optimize the OmniSci storage subsystem to take advantage of Intel® Optane™ DC persistent memory modules (Figure 6). Initial benchmarking results are extremely promising, allowing the possibility for OmniSci to support much larger datasets on a reduced hardware footprint.

Figure 6. Preliminary scaling results with OmniSciDB on Intel Optane DC persistent memory
We’re also exploring the use of oneAPI to target new Intel® hardware—in particular, GPUs with Xe Architecture from Intel, to take advantage of the OmniSci accelerated data-rendering pipeline. This allows the complete OmniSci stack, not just the OmniSciDB engine, to run on all Intel® platforms from the data center to the desktop or laptop.
Benefits to Data Scientists
Across the board, the collaboration provides significant benefits for data scientists. For the first time, they can run large-scale analytical computations over datasets with billions of rows of data on extremely efficient hardware footprints. This combination of scalability, performance, and familiarity makes the collaboration of Intel and OmniSci compelling as a high-performance data science environment to complement large-scale data processing and storage systems like data lakes and data warehouses.
Datasets up to a terabyte can be interactively analyzed and visualized on a laptop, and up to 10 TB or greater on a desktop-class system. The use of Arrow further promises the ability to seamlessly integrate machine-learning libraries such as Intel® Data Analytics Acceleration Library into the workflow.
Conclusion
OmniSci and Intel together provide a compelling new platform for data scientists. Integrating the capabilities of an open, hardware-aware, high-performance analytics engine with the power of the Intel® technology ecosystem has already demonstrated great benefits in scalability, performance, and flexibility for data science workflows.
You can download and try OmniSciDB on your Mac* or Linux* laptop running Docker* by following these instructions. The following are some more resources to help you learn about OmniSci and build data science workflows:
- Blog: Overview of OmniSci in Data Science
- On-demand Webinars of OmniSci in Data Science
- Overview of OmniSci Architecture by Todd Mostak
______
You May Also Like