OneJoin uses Data Parallel C++ (DPC++) to execute an edit similarity join algorithm across different compute systems, solving a key problem in reading large-scale DNA sequences

Challenge
The rapid growth of data generation and the need for enterprise and cloud providers to store it is driving research into novel storage media, such as DNA (Deoxyribonucleic Acid). The growth of data expansion at 60 percent yearly1 is outpacing the 10 percent to 30 percent annual2 areal density increase of traditional magnetic and silicon media (called the Kryder rate). Compared to traditional media, DNA has properties that make it interesting for archival or cold storage (see Figure 1).
- Extreme density: 455 exabytes can theoretically be stored in a gram of DNA, compared to 30 TB in an LTO-8 magnetic tape cartridge
- Reliability: DNA lasts thousands to hundreds of thousands of years in cold, dry environments, as illustrated by modern DNA extractions from ancient biological specimens
- No technology obsolescence: the density of DNA is fixed, and therefore does not undergo architectural changes due to technology advancements over time as other storage technologies do

- Low cost of in vitro replication
The DNA molecule is a double-helix structure with four nucleotides, adenine (A), cytosine (C), guanine (G), and thymine (T), arranged in paired sequences. DNA used for data storage is a single-stranded sequence of these nucleotides, also referred to as an oligonucleotide (oligo).
Using DNA as a digital storage medium requires mapping digital data into a sequence of oligos using an encoding algorithm3. Once encoded, the oligonucleotide sequences are used to synthesize DNA through a chemical process. Due to limitations in synthesis, each DNA strand cannot be longer than a few hundred nucleotides. Thus, digital data is stored using millions of oligos.
Data stored in these oligos is read back by sequencing the DNA molecules using next-generation sequencing (NGS) machines. NGS machines read DNA by duplicating DNA strands and reading them hundreds or thousands of times (called high coverage). However, sequencing introduces errors, and the repeated reads do not always exactly correspond with the original oligos. Thus, to decode data stored in DNA and identify the original source oligos, a read consensus procedure is performed to align similar sequences across the many reads and infer the original oligos based on similar reads. The inferred oligos are then passed to a decoder to recover the original data. Read consensus is a compute-intensive process, even when dealing with small datasets. It becomes an intractable challenge when dealing with lots of DNA and hundreds of millions of reads.

Figure 1. Emerging DNA storage technology offers highly condensed, large-scale cold data storage.
Solution
Dr. Raja Appuswamy is an assistant professor in the Data Science Department at Eurecom (see Figure 2). Eurecom is part of the OligoArchive project, an EU-funded initiative with a consortium of six partners. Professor Appuswamy studies how to build high-performance data management systems for enterprise OLTP, data warehousing, and data-intensive scientific domains like computational genomics. This requires working within several disciplines, such as databases, bioinformatics, storage systems, cloud computing, and high-performance computing.

Figure 2. Professor Raja Appuswamy and researcher Eugenio Marinelli.
“The main technical challenge in my research is developing scalable algorithms and systems that can effectively exploit modern hardware to reliably store and efficiently process data,” he said.
With the OligoArchive project, his team is working on efficient encoding and decoding algorithms for storing and retrieving structured databases in DNA.
“A typical NGS run today can easily produce hundreds of millions or billions of reads that need to be aligned to identify the original oligos,” Appuswamy explained. “But the algorithms traditionally used for this do not scale to large datasets. That’s because of intractable distance metrics (edit distance4) for comparing reads and the inability to utilize massive heterogeneous parallelism available on today’s servers.”
To address these challenges, he created the OneOligo project.
“In OneOligo, we are bringing together work from data management (edit similarity join processes using low-distortion embedding) and cross-architecture data parallelism using oneAPI to solve the read consensus problem for DNA storage,” he added.
The OneOligo project covers two phases. In the last five months constituting the first phase, Appuswamy and his colleague have developed OneJoin. OneJoin is a data-parallel, edit similarity join algorithm that exploits portable parallelism enabled by DPC++.
“We have a fully functional DPC++ implementation of OneJoin,” Appuswamy said. “We have tested our solution on a variety of processors including Intel® CPUs, Intel® HD Graphics Gen9 on Intel® DevCloud, and even NVIDIA RTX 2080 GPUs using the Codeplay LLVM backend. Our evaluation demonstrates that OneJoin can accelerate certain key tasks by over a factor of 100 (see Figure 3). It provides an overall 20x speedup over state-of-the-art join implementations, making it a scalable building block for the DNA read consensus problem.”
In the second phase of the project, they will build on OneJoin and implement a scalable read clustering solution, and then study various performance and accuracy tradeoffs in context of DNA storage.
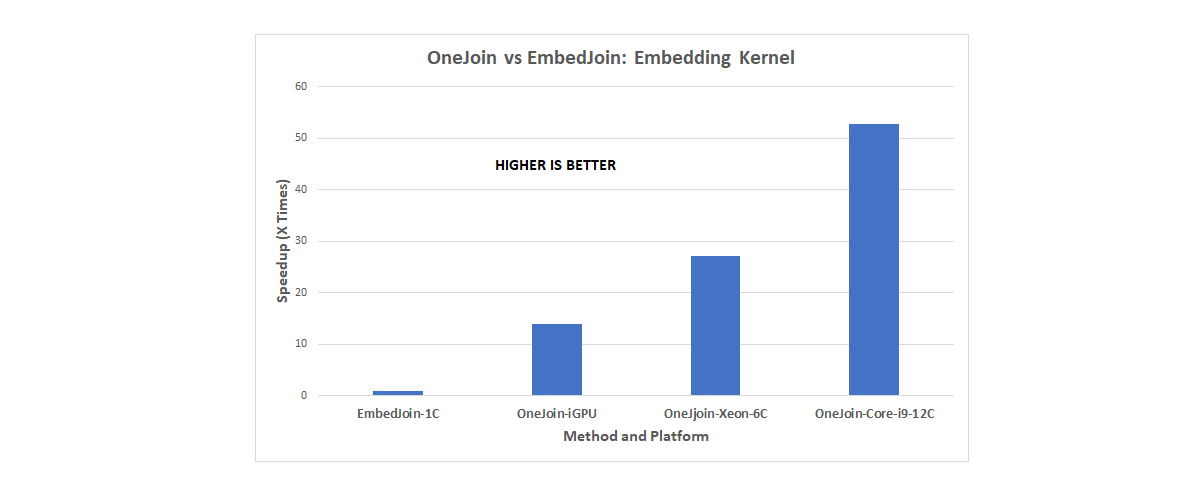 Figure 3. OneJoin speedup results (provided by Eurecom).
Figure 3. OneJoin speedup results (provided by Eurecom).
History
OneOligo is a scalable decoding algorithm that eliminates computational bottlenecks in the DNA data storage pipeline being developed within the OligoArchive project. OneOligo started with a single-threaded implementation of a join algorithm. The team profiled it with Intel® VTune™ Profiler to identify the most time-consuming stages and developed data-parallel kernels for those key stages. These steps formed the “fork” stages of a fork-join parallel execution model.
“We used DPC++ to implement the kernels. Doing so makes it possible to fork out our kernels across multiple architectures using a single code base,” said Appuswamy.
The join stages of the fork-join execution model are handled on the host; they involve memory management and data structure reorganization.
“We used the Intel® oneAPI DPC++ Library (oneDPL) and Intel® oneAPI Threading Building Blocks (oneTBB) to parallelize various data-structure-management tasks, like sorting and duplicate elimination,” Appuswamy continued. “Cross-architecture fork-join is an effective way to exploit CPUs and GPUs simultaneously. The use of different device selectors makes it possible to achieve this with ease,” he concluded.
The entire process takes less than 30 minutes (see Figure 4).

Figure 4. End-to-end experiment process (image courtesy of Dr. Raja Appuswamy).
The OneOligo read consensus method will be used by OligoArchive to decode data stored in DNA. Appuswamy will make the OneJoin source code publicly available and summarize their work in a research article to encourage research on cross-architecture data processing with oneAPI.
“The solutions we are developing have a much broader applicability,” he explained. “For instance, OneJoin’s edit similarity join can also be used for several other tasks in the data management domain, including data integration, data cleaning, entity recognition, and others. Similarly, the read clustering solution that will be developed can be used for multiple sequence alignment in computational genomics.”
Conclusion
OligoArchive’s vision is to overcome the biological and computational bottlenecks that hinder the adoption of DNA storage. Using oneAPI, OneJoin implements a key component of the decoding stack.
“To our knowledge, OneJoin is the only known edit similarity join algorithm that is both scalable and accurate for a variety of datasets,” Appuswamy stated. “We have been able to clearly demonstrate the benefit of cross-architecture portable parallelism offered by DPC++ and oneAPI for scalable data processing.”
“oneAPI enabled us to rapidly develop scalable, single-source, data-parallel algorithms for DNA data storage that can target CPUs and GPUs (integrated and discrete), using a unified programming model.”
Dr. Raja Appuswamy, Eurecom
“We chose DPC++ for two reasons,” concluded Appuswamy. “It made it easy to quickly develop a data-parallel implementation and test a preliminary prototype before diving into more detailed architecture-specific optimizations and because of its cross-architecture and cross-platform portability.”
Dr. Raja Appuswamy, Eurecom
Enabling Technologies
Dr. Raja Appuswamy used the following tools and technologies to help develop OneJoin.
Software:
- Intel® oneAPI Base Toolkit
- Intel® Parallel Studio XE
- Intel® VTune™ Profiler
- Intel® oneAPI Threading Building Blocks (oneTBB)
- Intel® oneAPI DPC++ Library (oneDPL)
Hardware:
- Intel® Xeon® E-2146G Processor
- Intel® Core™ i9-10920X Processor
- Intel® HD Graphics (Gen9)
- Intel® DevCloud
Resources and Recommendations
Footnotes
- Cold Storage in the Cloud: Trends, Challenges, and Solutions
- AIP Advances, 125222 (2019), 125222© 2019 Authors. Tape in the cloud—Technology developments and roadmaps supporting 80TB cartridge capacities
- Store Digital Data into DNA: A Comparative Study of Quaternary Code Construction
- Computing the edit-distance is a nontrivial computational problem because we must find the best alignment among exponentially many possibilities. For example, if both strings are 100 characters long, then there are more than 10^75 possible alignments. Dynamic Programming: Edit Distance